Post by: curiouscat on December 01, 2012, 08:39:17 AM
PS. The Preview feature doesn't seem to render Tex? Or maybe it's only my stupid browser?
Wondering what others think..
Post by: Borek on December 01, 2012, 08:55:16 AM
We can devote some thread to testing and delete the content now and then, no problem. I can't think of a better solution at the moment. But I am open to suggestions.
It is possible to create some simple sandbox for MathJax, but it would be not integrated with the forum itself, not the idea that I like.
Post by: curiouscat on December 01, 2012, 08:56:56 AM
We can devote some thread to testing and delete the content now and then, no problem.
That's the solution I had in mind.
Post by: Dan on December 01, 2012, 09:07:55 AM
You can PM yourself Tex to test it.
Post by: curiouscat on December 01, 2012, 09:10:21 AM
This would be useful, I also never get Tex right first time.Just had a thought, can you PM yourself to test it?
You can PM yourself Tex to test it.
Yep! Works.
Post by: curiouscat on December 12, 2012, 01:23:18 PM
http://www.chemicalforums.com/index.php?topic=64389.0;topicseen
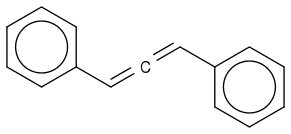
![c1ccccc1CC([Fe+][CO4])Cc2ccccc2](https://www.chemicalforums.com/SMILES/incorrect-SMILES.png)
Post by: Yggdrasil on December 12, 2012, 01:33:36 PM
http://www.forkosh.com/mimetextutorial.html
Post by: curiouscat on December 12, 2012, 01:36:21 PM
The following site has a nice sandbox for testing out LaTeX code (and is a good reference as well).
http://www.forkosh.com/mimetextutorial.html
Thanks! Loved the site!
Post by: Borek on December 12, 2012, 02:13:18 PM
Post by: Borek on December 12, 2012, 02:15:20 PM
Apparently SMILES don't like new lines in the SMILES code. Obvious, but I have never thought about it earlier.
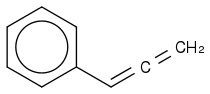
Post by: curiouscat on December 12, 2012, 02:16:58 PM
Apparently SMILES don't like new lines in the SMILES code. Obvious, but I have never thought about it earlier.
Ahhhh! :P Thanks!
Post by: Borek on December 13, 2012, 08:11:56 AM
ALT text is still wrong, but this is more difficult to deal with due to the way things are organized in the forum code.

Post by: curiouscat on December 13, 2012, 08:54:57 AM
Post by: sjb on December 13, 2012, 09:13:59 AM
Post by: Borek on December 13, 2012, 09:45:13 AM
Although rendered image is not following IUPAC suggestions - it should be more compact. There are things that can be improved in the indigo, so far one of my suggestions made its way into the code (but I have no idea when it is will be released).
Post by: curiouscat on December 13, 2012, 09:58:41 AM
Not always good for forum formatting, but apparently the engine survived.
Although rendered image is not following IUPAC suggestions - it should be more compact. There are things that can be improved in the indigo, so far one of my suggestions made its way into the code (but I have no idea when it is will be released).
Still impressive. I was expecting an error message when I put that in. :)
Post by: curiouscat on December 13, 2012, 12:24:39 PM
Although rendered image is not following IUPAC suggestions - it should be more compact.
What are IUPAC conventions on image rendering? I am curious. Do you have a link? I couldn't google anything up.
Post by: Borek on December 13, 2012, 12:36:35 PM
Post by: curiouscat on January 02, 2013, 03:55:58 AM
Buckministerfullerene
Stealing the SMILES string off ChemSpider / Wikipedia:
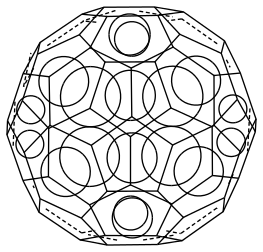
Incidently shouldn't the molecule have 60 C entities since it's a C60? Was surprised that the Wikipedia / Chemspider strings only went up to C11. What gives?
Even the smaller Dodecahedrane doesn't seem to work.
PS. As an aside Annulene looks pretty
Though this might have been a better way to render it (also as per Borek's very nice IUPAC guidelines):
(https://www.chemicalforums.com/proxy.php?request=http%3A%2F%2Fupload.wikimedia.org%2Fwikipedia%2Fcommons%2Fthumb%2F3%2F33%2F%252818%2529Annulene.svg%2F166px-%252818%2529Annulene.svg.png&hash=377fc6e285ebd6dbc5f44d8b9e59ebf97353858f)
Post by: Borek on January 02, 2013, 04:42:19 AM
c12c3c4c5c1c6c7c8c2c9c1c3c2c3c4c4c%10c5c5c6c6c7c7c%11c8c9c8c9c1c2c1c2c3c4c3c4c%10c5c5c6c6c7c7c%11c8c8c9c1c1c2c3c2c4c5c6c3c7c8c1c23
Incidently shouldn't the molecule have 60 C entities since it's a C60? Was surprised that the Wikipedia / Chemspider strings only went up to C11. What gives?
To quote Daylight SMILES description:
Quote
If desired, digits denoting ring closures can be reused.
And apparently that's what is done here.
Quote
Though this might have been a better way to render it (also as per Borek's very nice IUPAC guidelines):
(https://www.chemicalforums.com/proxy.php?request=http%3A%2F%2Fupload.wikimedia.org%2Fwikipedia%2Fcommons%2Fthumb%2F3%2F33%2F%252818%2529Annulene.svg%2F166px-%252818%2529Annulene.svg.png&hash=377fc6e285ebd6dbc5f44d8b9e59ebf97353858f)
Forwarding to indigo team as well.
Post by: curiouscat on January 02, 2013, 04:49:44 AM
All I can do is to forward these problems to the Indigo team (and be sure I will, although I know they are having a time off right now).
Sure. Thanks!
Just doing our lil' bit to make the Open Source Ecosystem a wee bit better, one bug report at a time. :)
Quote
If desired, digits denoting ring closures can be reused.
And apparently that's what is done here.
Of course. Makes sense. Thanks! Somehow I was expecting to see a C60 index; dumb in hindsight. Lost sight of what these indexes meant.
Post by: curiouscat on January 22, 2013, 06:02:51 AM
You can PM yourself Tex to test it.
"You have exceeded the limit of 10 personal messages per hour."
Not a big deal at all, but just was funny how one can keep hitting the law of unintended consequences. ;D
Who'd have thought of Dan's idea and then all because it took stupid me 10 attempts to get this SMILES right:
![[*]C1OC1[*]](https://www.chemicalforums.com/SMILES/5f1040394a06207d5431.png)
Was even funnier coz' Reply or Quote is the only way to get the SMILES in the past message sandbox attempts and SMF won't let me try that now. (Ha! If someone gets what I write! ).
Guess I'll be in the doghouse for an hour! :P
PS. Is there a way to get R1 and R2 rendered in SMILES?
Post by: Borek on January 22, 2013, 06:07:40 AM
![[*:1]C1OC1[*:2]](https://www.chemicalforums.com/SMILES/ead3eec716db62825d96.png)
Post by: curiouscat on January 22, 2013, 06:13:50 AM
Ah! Thanks! :-[
![[*:1]C(Cl)C(O)[*:2]>>[*:1]C1OC1[*:2]](https://www.chemicalforums.com/SMILES/56e6a3019bf5ecab8a93.png)
Looks a bit ugly here... ???
Post by: Borek on January 22, 2013, 06:26:28 AM
Looks a bit ugly here... ???
Another thing to report, once I am sure it is something to report :-\
Turned out problems with C60 are somewhere on our side - I was unlucky enough to hit two separate bugs on the forum and on my own computer, I assumed if it doesn't work here nor there it must be a problem with the SMILES renderer - but that's not the case.
Post by: curiouscat on January 22, 2013, 06:29:40 AM
Another thing to report, once I am sure it is something to report :-\
Turned out problems with C60 are somewhere on our side - I was unlucky enough to hit two separate bugs on the forum and on my own computer, I assumed if it doesn't work here nor there it must be a problem with the SMILES renderer - but that's not the case.
Did you have to do a lot of coding to integrate SMILES on "our side"? Wondering what "our side" bugs are like..
Post by: Borek on January 22, 2013, 07:10:01 AM
Did you have to do a lot of coding to integrate SMILES on "our side"? Wondering what "our side" bugs are like..
Something like several hours to modify forum sources - including time needed to find out how the BBC tags are analyzed and where. But it took several days to compile the renderer, as we discussed in the past.
And I have no idea what the bug is - yet. Most likely % sign is treated incorrectly. Trick is, I don't pass it - I have modified the program so that what is passed is just a hexadecimal representation of the original SMILES (so 434F is passed instead of CO), this way I don't have to care about the data sanitization (assuming the renderer just ignores incorrect input; I bet it does).
Post by: curiouscat on January 22, 2013, 07:11:53 AM
Did you have to do a lot of coding to integrate SMILES on "our side"? Wondering what "our side" bugs are like..
Something like several hours to modify forum sources - including time needed to find out how the BBC tags are analyzed and where. But it took several days to compile the renderer, as we discussed in the past.
And I have no idea what the bug is - yet. Most likely % sign is treated incorrectly. Trick is, I don't pass it - I have modified the program so that what is passed is just a hexadecimal representation of the original SMILES (so 434F is passed instead of CO), this way I don't have to care about the data sanitization (assuming the renderer just ignores incorrect input; I bet it does).
Wow, sounds crazy difficult. Somehow I was thinking it's a "plugin". :P
Post by: Borek on February 09, 2013, 08:57:39 AM
Was trying to throw Fullerenes at SMILES for fun but can't seem to make it work. Any ideas?
Buckministerfullerene
Stealing the SMILES string off ChemSpider / Wikipedia:
Works now. Stupid bug.
Quote
Works, but I would not call it pretty :o
Post by: curiouscat on February 09, 2013, 10:26:15 AM
Post by: Dan on February 19, 2013, 06:12:57 AM
![C[C@H]1CC(=O)[C@H](C)CC1=O](https://www.chemicalforums.com/SMILES/b0c6acf2d406385521f0.png)
Claims to be chiral, but is in fact a meso compound (achiral).
Same is true for the rendering of ribitol:
[C@H](O)[C@H](O)CO](https://www.chemicalforums.com/SMILES/b46951fa0ca4758e1c90.png)
Same problem here:
![C[C@H]1CCCC[C@H]1C](https://www.chemicalforums.com/SMILES/9fde79e40f235b9d6bbe.png)
Post by: Borek on February 19, 2013, 06:42:31 AM
Post by: sjb on February 19, 2013, 07:46:42 AM
Post by: Borek on February 19, 2013, 07:51:13 AM
and
Hm, when chirality doesn't matter isn't it better to just show the molecule as
 ?
?Post by: sjb on February 19, 2013, 07:58:01 AM
Hm, when chirality doesn't matter isn't it better to just show the molecule as?
Not really, in this case there are a cis pair and a trans pair of diasteromers.
Your compound also includes
![C(C)(C)[C@H]1CC[C@H](C1)O](https://www.chemicalforums.com/SMILES/6e4f5b4d73e9a15f55f6.png) and
and ![C(C)(C)[C@@H]1CC[C@@H](C1)O](https://www.chemicalforums.com/SMILES/0c7828cb353ae11cee01.png) to my mind.
to my mind.Post by: Borek on February 19, 2013, 08:55:24 AM
I mean - I see why the "flat" representation can be too wide (mean to many things), but I have no idea how the racemic mixture SHOULD be represented?
Post by: sjb on February 19, 2013, 09:09:16 AM
The cases we are discussing are ~ slide 20-23
I seem to recall that in the GSK system (as referenced above) there were "chiral" and "racemic" flags you could put on the molecule when imported from ISIS draw (as was) which helped distinguish these cases. Not sure how it worked on the back-end though :(
Post by: Dan on February 19, 2013, 10:24:33 AM
I have no idea how the racemic mixture SHOULD be represented?
I would draw one enantiomer and indicate that it is racemic with a (±) symbol underneath it.
Post by: Dan on February 20, 2013, 01:47:49 PM
![C[C@H]1CCCCC1](https://www.chemicalforums.com/SMILES/ff9ff5204d44f4decab5.png)
Post by: billnotgatez on March 03, 2013, 05:37:45 PM
or
2C2H2 + 5O2 :rarrow: 4CO2 + 2H2O
I wrote the above two formulas using the formatting available in the reply box
Question 1 - which one do you like
Question 2 - could I have used LATEX or SMILES
Post by: Borek on March 03, 2013, 06:12:49 PM
In this particular case I would not use neither SILES nor LaTeX. Call it a personal preference.
Post by: billnotgatez on March 03, 2013, 06:20:27 PM
but
Do we have links to teaching
SMILES and LaTex handy
I think I am being common sensed challenged here, since I am not seeing any
Post by: Borek on March 03, 2013, 06:47:37 PM
Post by: billnotgatez on March 03, 2013, 07:55:00 PM
SMILES at Daylight site
http://www.daylight.com/dayhtml/doc/theory/theory.smiles.html
SMILES at wikipedia
http://en.wikipedia.org/wiki/SMILES
ChemSketch
http://www.acdlabs.com/download/
LaTeX
http://en.wikibooks.org/wiki/LaTeX/Mathematics)
Post by: curiouscat on March 03, 2013, 10:49:43 PM
In this particular case I would not use neither SILES nor LaTeX. Call it a personal preference.
+1 SMILES only becomes relevant above a certain minimum complexity of molecule. Usually, no one needs to be reminded how, say, ethane is structured internally.
Post by: Borek on March 04, 2013, 04:04:18 AM
Did I miss any
SMILES at Daylight site
http://www.daylight.com/dayhtml/doc/theory/theory.smiles.html
SMILES at wikipedia
http://en.wikipedia.org/wiki/SMILES
ChemSketch
http://www.acdlabs.com/download/
LaTeX
http://en.wikibooks.org/wiki/LaTeX/Mathematics)
I believe it is a good starting collection, at least for the forum. There is much more to LaTeX, but most of it is of no use for us.
Post by: billnotgatez on March 21, 2014, 07:37:43 AM
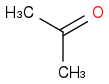
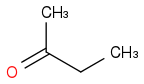 or
or 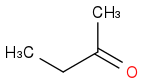
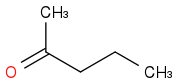
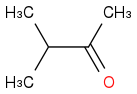
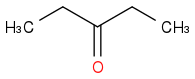
Just smiles testing some acids
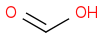
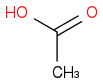
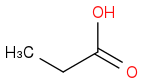
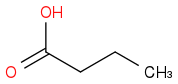 or
or 
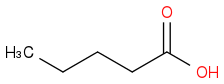
Post by: billnotgatez on April 26, 2014, 03:30:54 AM
[tex] \frac {(2)}{(4)} = \frac {(1)}{(2)} [/tex]
[tex] \frac {(P_1 . V_1)}{(n_1 . T_1)} = \frac {(P_2 . V_2)}{(n_2 . T_2)} [/tex]
Post by: Xenonman on April 29, 2014, 10:37:18 PM
Zn+2(ac) + Y-4(ac) :rarrow: [ZnY]-2(ac)
nZn+2=nY-4= 0.0011 mol
Don't mind me. Just me failing at making divisions in tex.
Yes, I wrote states as subscripts. That's the way it's done around here.
Post by: Borek on April 30, 2014, 02:32:00 AM
Yes, I wrote states as subscripts. That's the way it's done around here.
There is no such thing as "here" here.
IUPAC rules are clear and they are defined to be used by everyone.
Post by: curiouscat on April 30, 2014, 03:02:23 AM
Yes, I wrote states as subscripts. That's the way it's done around here.
Around here is where?
Post by: curiouscat on April 30, 2014, 03:03:32 AM
IUPAC rules are clear and they are defined to be used by everyone.
Yes the rules are clear but how does one mandate everyone to use them? :)
Post by: Borek on April 30, 2014, 04:59:25 AM
IUPAC rules are clear and they are defined to be used by everyone.
Yes the rules are clear but how does one mandate everyone to use them? :)
Technically you don't, but it is a matter of being clear and unambiguous. What is VO2+? IUPAC rules tell you how to write it in an unambiguous way, why not stick to them?
Post by: curiouscat on April 30, 2014, 08:27:16 AM
Technically you don't, but it is a matter of being clear and unambiguous. What is VO2+? IUPAC rules tell you how to write it in an unambiguous way, why not stick to them?
Agreed.
Post by: billnotgatez on April 30, 2014, 08:40:58 AM
I did a quick GOOGLE and did not find this one
I have yet to peruse the IUPAC rules
Post by: Borek on April 30, 2014, 09:15:06 AM
http://www.chemicalforums.com/index.php?topic=8373.msg38043#msg38043
Post by: Xenonman on April 30, 2014, 10:01:44 AM
Those zinc equations were for an assignment. In Spanish again.
I was taught to use (ac), as you can see. This is what every future Olympic will be taught, so I'll try to show this discrepancy, and an example of what happens when they use (ac) instead of (aq). I'll use (aq) as long as I don't forget when posting here, also.
Post by: billnotgatez on May 03, 2014, 10:26:10 PM
Cobalt(II) oxide
WIKI
![[Co]=O](https://www.chemicalforums.com/SMILES/239448b62db7e5787575.png)
SPIDER
![[O-2].[Co+2]](https://www.chemicalforums.com/SMILES/478055b88c05730445cb.png)
Cobalt(III) oxide
SPIDER
![O=[Co]O[Co]=O](https://www.chemicalforums.com/SMILES/f8ed263935734d0cb1e9.png)
Cobalt(II,III) oxide
WIKI
![[Co]=O.O=[Co]O[Co]=O](https://www.chemicalforums.com/SMILES/ef77c24915aa8e4cd0b3.png)
SPIDER
![[Co+2].[Co+2].[Co+2].[O-2].[O-2].[O-2].[O-2]](https://www.chemicalforums.com/SMILES/41d70cabdfe92aae2bf1.png)
Cobalt(II) carbonate
SPIDER
![C(=O)([O-])[O-].[Co+2]](https://www.chemicalforums.com/SMILES/b3c2ffd6c83ccc4b783c.png)
Post by: billnotgatez on May 04, 2014, 05:26:43 AM
Glycerol or Glycerin C3H8O3
WIKI
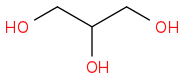
SPIDER

Post by: Borek on May 04, 2014, 06:41:14 AM
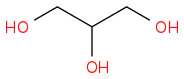
Post by: curiouscat on May 04, 2014, 08:59:28 AM
Strange. Why C(C(CO)O)O when OCC(O)CO is much simpler? Perhaps wiki has some computer generated code.
Yes indeed. That convoluted version is almost impossible to create by hand accidentally.
Post by: billnotgatez on May 04, 2014, 09:22:35 AM
When I looked up the smiles for the Cobalt oxides WIKI was different than SPIDER
But they both had the same for the Glycerin (Glycerol)
Is there a source for "standard smiles" compounds?
Post by: Borek on May 04, 2014, 02:50:01 PM
IOW: SMILES are a good equivalent of stick and ball models, but not so good for infinite crystals.
Post by: billnotgatez on May 04, 2014, 10:25:10 PM
Quote
IOW: SMILES are a good equivalent of stick and ball models, but not so good for infinite crystals.Somewhere deep in my cranial cavity a light bulb is starting to illuminate.
Post by: Xenonman on May 13, 2014, 05:39:34 PM
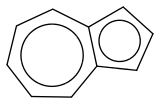 <- naphthalene attempt #1
<- naphthalene attempt #1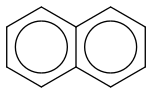 ;D
;D
![C#[C-][Na+]](https://www.chemicalforums.com/SMILES/67b1ab29942ba4a95d2d.png) What's with that carbon charge? o-o
What's with that carbon charge? o-oPost by: curiouscat on May 13, 2014, 11:08:57 PM
What's with that carbon charge? o-o
You wanted Sodium acetylide?
![C#C[Na]](https://www.chemicalforums.com/SMILES/75ef9a71e340f2686a00.png)
Post by: Borek on May 14, 2014, 02:36:20 AM
Post by: billnotgatez on May 27, 2014, 05:45:54 AM
I used
+ Additional Options...
Post by: billnotgatez on May 27, 2014, 06:04:40 AM
I used
+ Additional Options...
Post by: Xenonman on May 27, 2014, 08:22:17 PM
I can't believe it actually worked o.o
I'll go get a cookie.
Post by: billnotgatez on July 16, 2014, 06:19:36 AM
Post by: billnotgatez on August 20, 2014, 01:35:51 AM

Hexadecane

1-hexadecanol

hexadecanal

hexadecanoic acid
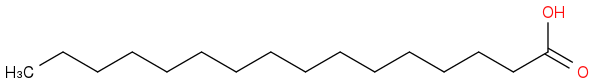
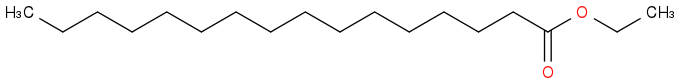
Ethyl hexadecanoate
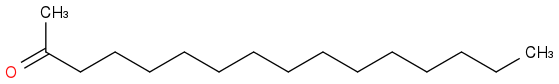
2-HEXADECANONE
Post by: curiouscat on August 20, 2014, 02:41:40 AM
Just idly wondering whether it will bend or scroll >:D

Post by: curiouscat on August 20, 2014, 02:43:11 AM


Post by: curiouscat on August 20, 2014, 02:44:18 AM



Post by: Dan on January 04, 2015, 10:41:42 AM
Any solution to this problem? [/list]
Post by: curiouscat on January 04, 2015, 10:53:20 AM
I can't have Li in smiles because the forum automatically converts [ Li ] (without spaces) to [list][li].
Any solution to this problem? [/list]
I thought [nobbc] [/nobbc] tags solved this. At least for Latex. But can't get it to work just now. Headscratch.
Post by: Borek on January 04, 2015, 04:03:58 PM
Post by: billnotgatez on July 06, 2015, 02:42:47 AM
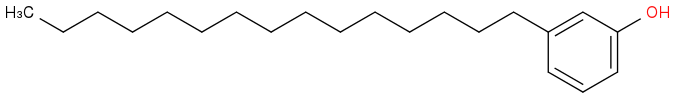
3-pentadecylphenol
http://pubchem.ncbi.nlm.nih.gov/compound/3-Pentadecylphenol#section=Top
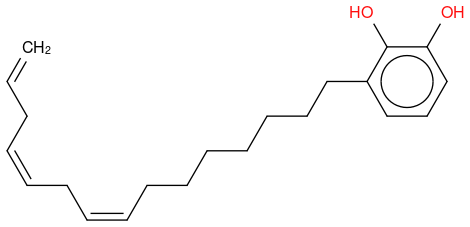
URUSHIOL (15:3) CAS: 83543-37-7 Formula:C21 H30 O2 MolWeight:314.466
http://www.chemcd.com/product/CCD02755975.html
Post by: curiouscat on July 06, 2015, 07:56:11 AM
3-pentadecylphenol
http://pubchem.ncbi.nlm.nih.gov/compound/3-Pentadecylphenol#section=Top
URUSHIOL (15:3) CAS: 83543-37-7 Formula:C21 H30 O2 MolWeight:314.466
http://www.chemcd.com/product/CCD02755975.html
Wonder how nature synthesizes these molecules. What pathways I mean.
Post by: billnotgatez on July 07, 2015, 03:26:51 AM
Quote
Wonder how nature synthesizes these molecules. What pathways I mean.I would be itching to find out as well (pun intended) >:D
more testing
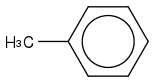
Toluene
https://en.wikipedia.org/wiki/Toluene
Post by: sjb on July 07, 2015, 04:05:23 AM
Post by: billnotgatez on July 18, 2015, 06:44:18 AM
(C5O2H8)n
C(=O)OC](https://www.chemicalforums.com/SMILES/87d3728ecf48bf3624b6.png)
https://en.wikipedia.org/wiki/Poly(methyl_methacrylate)
I copied the smiles from WIKI
How do you do the n in smiles?
Why is there a dot next to the C
More playing
Eugenol
C10H12O2
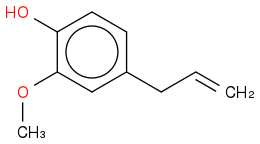
https://en.wikipedia.org/wiki/Eugenol
Post by: billnotgatez on August 06, 2015, 01:29:20 PM
which leads to an inaccurate link
so I am playing to see why
https://en.wikipedia.org/wiki/Poly(methyl_methacrylate) (https://en.wikipedia.org/wiki/Poly(methyl_methacrylate))
I noticed I had to use the insert hyperlink function to get the required result
It seems that just a copy paste does not always work for links
I think I noticed this before but probably did not remember and I do not remember why this is caused
Post by: Borek on August 06, 2015, 05:13:51 PM
Post by: billnotgatez on November 01, 2015, 11:25:01 PM
CAS Registry Number 112-80-1
Chemical formula C18H34O2
Molar mass 282.47 g·mol−1
formula CH3(CH2)7CH=CH(CH2)7COOH
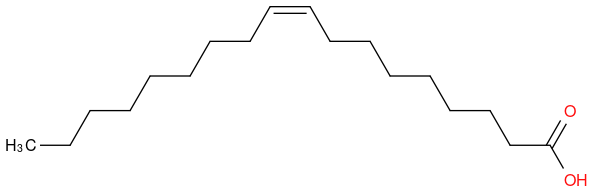
https://en.wikipedia.org/wiki/Oleic_acid (https://en.wikipedia.org/wiki/Oleic_acid)
Post by: swintarka on December 06, 2015, 11:56:33 AM
![C12=CC=CC(C3=CC=CC=C3)=C1CC=C2>[H][H]>C45CCC[C@H](C6CCCCC6)C4CCC5](https://www.chemicalforums.com/SMILES/df09ef5d2e1d757c4238.png)
Post by: billnotgatez on December 12, 2015, 02:44:55 AM
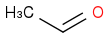
CH3CHO
Acetonitrile
CH3CN
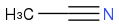
Post by: Doc.AElstein on January 04, 2016, 09:02:44 AM
Just practicing png’s. No Reply needed.
Thanks
Alan
( ThingsList ) ( Using PhotoBucket )
IMG
(https://www.chemicalforums.com/proxy.php?request=http%3A%2F%2Fi1065.photobucket.com%2Falbums%2Fu400%2FDocAElstein%2FThingsListTransposedSimplified_zps7daaxnag.jpg&hash=cee35d5e02005b51b79d2af22b290fade041094c) (http://s1065.photobucket.com/user/DocAElstein/media/ThingsListTransposedSimplified_zps7daaxnag.jpg.html)
.
Things table
(https://www.chemicalforums.com/proxy.php?request=http%3A%2F%2Fi1065.photobucket.com%2Falbums%2Fu400%2FDocAElstein%2FThingsTable1_zpsjtoml1hr.jpg&hash=7152e8e050809bede8b39002795243b65b64fce4) (http://s1065.photobucket.com/user/DocAElstein/media/ThingsTable1_zpsjtoml1hr.jpg.html)
(https://www.chemicalforums.com/proxy.php?request=http%3A%2F%2Fi1065.photobucket.com%2Falbums%2Fu400%2FDocAElstein%2FThingsTable2_zpsqrvhcpxm.jpg&hash=18da8487c710a258f095439ded85261442e0738a) (http://s1065.photobucket.com/user/DocAElstein/media/ThingsTable2_zpsqrvhcpxm.jpg.html)
(https://www.chemicalforums.com/proxy.php?request=http%3A%2F%2Fi1065.photobucket.com%2Falbums%2Fu400%2FDocAElstein%2FThingsTable3_zpsfsnhvyy1.jpg&hash=42a8c113b3945b85e01b0dbae6b3302826fa7580) (http://s1065.photobucket.com/user/DocAElstein/media/ThingsTable3_zpsfsnhvyy1.jpg.html)
(https://www.chemicalforums.com/proxy.php?request=http%3A%2F%2Fi1065.photobucket.com%2Falbums%2Fu400%2FDocAElstein%2FThingsTable4_zps5158wn8b.jpg&hash=be2141b20bf772e09e4d37c6eca8cca46a34183a) (http://s1065.photobucket.com/user/DocAElstein/media/ThingsTable4_zps5158wn8b.jpg.html)
……………………………………………..
Pro 2016-01-04
(https://www.chemicalforums.com/proxy.php?request=http%3A%2F%2Fi1065.photobucket.com%2Falbums%2Fu400%2FDocAElstein%2FPro2016_zpssprs0bqx.jpg&hash=66094f8256334124b8b2868756d5636d5ac37881) (http://s1065.photobucket.com/user/DocAElstein/media/Pro2016_zpssprs0bqx.jpg.html)
Try a Forum attatchment::-
http://www.chemicalforums.com/index.php?topic=69802.msg251351#msg251351
Post by: Doc.AElstein on January 07, 2016, 09:57:43 AM
Just Practicing Text File Uploading
No Reply Needed.
Thanks
Alan Elston
Box File Sharing first...
https://app.box.com/s/f7if1ry5i8tcgq3yq96lwljn23bnvnla
_ ...
Then try the
+ Addirional Options
( as per http://www.chemicalforums.com/index.php?topic=69802.msg251351#msg251351 )
Post by: billnotgatez on January 13, 2016, 09:55:28 PM
C6H4(NH2)2
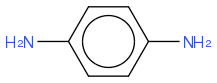
https://en.wikipedia.org/wiki/P-Phenylenediamine
m-Phenylenediamine
C6H4(NH2)2
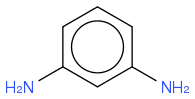
https://en.wikipedia.org/wiki/M-Phenylenediamine
o-Phenylenediamine
C6H4(NH2)2
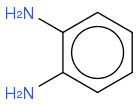
https://en.wikipedia.org/wiki/O-Phenylenediamine
Indophenol
C12H9NO2 or OC6H4NC6H4OH
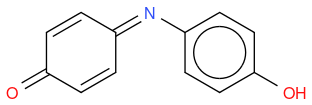
https://en.wikipedia.org/wiki/Indophenol
Post by: Doc.AElstein on January 14, 2016, 07:07:35 PM
(https://www.chemicalforums.com/proxy.php?request=http%3A%2F%2Fi1065.photobucket.com%2Falbums%2Fu400%2FDocAElstein%2Fcarotenoids_zpsv73equ0r.jpg&hash=20de5229ba35a14abc64a7649eedef777be1afeb) (http://s1065.photobucket.com/user/DocAElstein/media/carotenoids_zpsv73equ0r.jpg.html)
(https://www.chemicalforums.com/proxy.php?request=http%3A%2F%2Fi1065.photobucket.com%2Falbums%2Fu400%2FDocAElstein%2Fcarotenoids2_zpskvfxanii.jpg&hash=c0688f261feb659432d0b2ee3720aec54ce26d0d) (http://s1065.photobucket.com/user/DocAElstein/media/carotenoids2_zpskvfxanii.jpg.html)
Post by: Doc.AElstein on January 14, 2016, 08:36:32 PM
(https://www.chemicalforums.com/proxy.php?request=http%3A%2F%2Fi1065.photobucket.com%2Falbums%2Fu400%2FDocAElstein%2Fbeta-alanin_zpsbgbzpowf.jpg&hash=dcf93efd4efb7fcfe40473f234ae7c4b26d193b8) (http://s1065.photobucket.com/user/DocAElstein/media/beta-alanin_zpsbgbzpowf.jpg.html)
(https://www.chemicalforums.com/proxy.php?request=http%3A%2F%2Fi1065.photobucket.com%2Falbums%2Fu400%2FDocAElstein%2Fbeta-alanin2_zpsj3na91zs.jpg&hash=dfe2368842a11ae2745989dbcae73a1085155201) (http://s1065.photobucket.com/user/DocAElstein/media/beta-alanin2_zpsj3na91zs.jpg.html)
Post by: billnotgatez on March 22, 2016, 10:01:50 AM
C5H12
Post by: Doc.AElstein on April 29, 2016, 01:32:40 PM
| This came from clcking on an icon above |
_....
This came from a BB Code Generator Code: that I have
Using Excel 2007
[Table="width:, class:grid"]
| This came | from a |
| BB Code | Generting |
| Code I have | Working from |
| Office | Excel |
| Leith2 |
[color=lightgrey]Using Excel 2007[/color]
[size=1][Table="width:, class:grid"][table][tr][td]This came[/td][td]from a [/td][/tr]
[tr][td]BB Code[/td][td]Generting[/td][/tr]
[tr][td]Code I have[/td][td]Working from[/td][/tr]
[tr][td]Office[/td][td]Excel[/td][/tr]
[/table][/size][Table="width:, class:grid"][table][tr][td][b]Leith2[/b][/td][/tr][/table]
OK, need to chop some bits from the BB Code sting in the Code..
then...
Using Excel 2007
| This came | from a |
| BB Code | Generting |
| Code I have | Working from |
| Office | Excel |
Sucess ! :)
[color=lightgrey]Using Excel 2007[/color]
[size=1][table][tr][td]This came[/td][td]from a [/td][/tr]
[tr][td]BB Code[/td][td]Generting[/td][/tr]
[tr][td]Code I have[/td][td]Working from[/td][/tr]
[tr][td]Office[/td][td]Excel[/td][/tr]
[/table]
..
FYI. ... "no parse stuff! "
[noparse][color=lightgrey]Using Excel 2007[/color]
[size=1][table][tr][td]This came[/td][td]from a [/td][/tr]
[tr][td]BB Code[/td][td]Generting[/td][/tr]
[tr][td]Code I have[/td][td]Working from[/td][/tr]
[tr][td]Office[/td][td]Excel[/td][/tr]
[/table][/noparse]
Using Excel 2007
| This came |
| BB Code |
Using Excel 2007
| [td="bgcolor:#FFFF00"]This came |
| [td="bgcolor:#FFC000"]BB Code |
_..
copy from forum to word then back
Using Excel 2007
This came
BB CodeUsing
_..
copy from forum to Excel then copy back
Using Excel 2007
This came
BB Code
_....
Direct copy from Excel
This came from a
BB Code Generting
Code I have Working from
Office Excel
_....
Modified BB Code looks best
:)
Using Excel 2007
| This came | from a |
| Modified BB Code | Generting |
| Code I have | Working from |
| Office | Excel |
Post by: Doc.AElstein on April 29, 2016, 05:46:10 PM
| Per 100g | per 12.3g | per 17.5g | |
| Kcal | 350 | 59.5 | 43.05 |
| Fat | 0.3 | 0.051 | 0.0369 |
| Protein | 85 | 14.45 | 10.455 |
| Carbohydrates | 0.1 | 0.017 | 0.0123 |
| Sugars | 0.1 | 0.017 | 0.0123 |
| Water | 11 | 1.87 | 1.353 |
| Total dietary fibre | 0 | 0 | 0 |
| Cholesterol | 0 | 0 | 0 |
| Mineral | 1.7 | 0.289 | 0.2091 |
| Vitamin B6 | 0.00001 | 0.0000017 | 0.00000123 |
| Niacinequivalent | 0.0001 | 0.000017 | 0.0000123 |
| Sodium | 0.06 | 0.0102 | 0.00738 |
| Potassium | 0.07 | 0.0119 | 0.00861 |
| Magnesium | 0.011 | 0.00187 | 0.001353 |
| Copper | 0.00005 | 0.0000085 | 0.00000615 |
| Zinc | 0.0002 | 0.000034 | 0.0000246 |
| Chloride | 0.1 | 0.017 | 0.0123 |
| Fluoride | 0.00003 | 0.0000051 | 0.00000369 |
| Iodide | 0.000006 | 0.00000102 | 7.38E-07 |
| Manganese | 0.00004 | 0.0000068 | 0.00000492 |
| Sulphur | 0.1 | 0.017 | 0.0123 |
| Arginine | 7.8 | 1.326 | 0.9594 |
| Cystin | 0 | 0 | 0 |
| Histidine | 0.64 | 0.1088 | 0.07872 |
| Isoleucine | 1.45 | 0.2465 | 0.17835 |
| Leucine | 3.6 | 0.612 | 0.4428 |
| Lysine | 3.95 | 0.6715 | 0.48585 |
| Methionine | 0.5 | 0.085 | 0.0615 |
| Phenylalanine | 2.1 | 0.357 | 0.2583 |
| Threonine | 1.9 | 0.323 | 0.2337 |
| Tryptophane | 0.003 | 0.00051 | 0.000369 |
| Tyrosine | 0.27 | 0.0459 | 0.03321 |
| Valine | 2.45 | 0.4165 | 0.30135 |
| Alanine | 10.5 | 1.785 | 1.2915 |
| Aspartic acid | 6.4 | 1.088 | 0.7872 |
| Glutamic acid | 10.5 | 1.785 | 1.2915 |
| Glycine | 23.9 | 4.063 | 2.9397 |
| Proline | 14.2 | 2.414 | 1.7466 |
| Serine | 3.65 | 0.6205 | 0.44895 |
| essent. amino acids | 22 | 3.74 | 2.706 |
| nonessent. amino acids | 63.8 | 10.846 | 7.8474 |
Post by: Arkcon on May 01, 2016, 03:31:42 PM
![[O-]S(=O)(=O)[O-].[O-]S(=O)(=O)[O-].[Al+3].[K+]](https://www.chemicalforums.com/SMILES/0d24e8cb1481e8200de7.png)
Alum. Hum. I don't know what I was expecting from an inorganic SMILES, but that just doesn't seem worth the trouble under any circumstances.
([O-])[O-].[Ag+]](https://www.chemicalforums.com/SMILES/4b2e2d1a4ccbc89d6b2f.png)
Post by: billnotgatez on May 02, 2016, 05:51:52 PM
CO(NH2)2
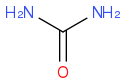
https://en.wikipedia.org/wiki/Urea
Post by: Doc.AElstein on May 04, 2016, 08:15:37 PM
This ...
Using Excel 2007
|
came from this:
[color=lightgrey]Using Excel 2007[/color]
[table][tr][td]
[table][tr][td][table][tr][td][CENTER][COLOR=#A5A5A5]Grey[/COLOR][/CENTER][/td][/tr][/table][table][tr][td][CENTER][/CENTER][/td][/tr]
[/table]
[/td][/tr][/table][/td][/tr][/table]
Post by: billnotgatez on May 10, 2016, 10:18:13 PM
C3H8O or C3H7OH or CH3CHOHCH3 or i-PrOH or (CH3)2CHOH
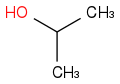
https://en.wikipedia.org/wiki/Isopropyl_alcohol
---------------------------------------------------
acetone or propanone or propan-2-one
C3H6O or (CH3)2CO
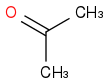
https://en.wikipedia.org/wiki/Acetone
---------------------------------------------------
Post by: billnotgatez on October 08, 2016, 01:42:50 PM
Post by: sjb on November 03, 2016, 05:49:00 AM
![Cl[C@H]1C[C@H]2CC[C@@H]1C2](https://www.chemicalforums.com/SMILES/a113521ea23ee233b40c.png)
Post by: billnotgatez on November 24, 2016, 11:03:44 AM
propane-1,2-diol
C3H8O2
Molar mass 76.10 g/mol
Density 1.036 g/cm3
CAS Number 57-55-6
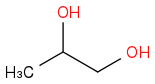
https://en.wikipedia.org/wiki/Propylene_glycol (https://en.wikipedia.org/wiki/Propylene_glycol)
Glycerol
glycerine or glycerin or 1,2,3-Propanetriol or 1,2,3-Trihydroxypropane
C3H8O3
Molar mass 92.09 g/mol
Density 1.261 g/cm3
CAS Number 56-81-5

https://en.wikipedia.org/wiki/Glycerol (https://en.wikipedia.org/wiki/Glycerol)
Ethanol
ethyl alcohol
C2H6O or CH3CH2OH or C2H5OH or EtOH
Molar mass 46.068 g/mol
Density 0.7893 g/cm3 (at 20 °C)
CAS Number 64-17-5
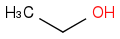
https://en.wikipedia.org/wiki/Ethanol (https://en.wikipedia.org/wiki/Ethanol)
Post by: billnotgatez on January 31, 2017, 06:18:23 PM
https://en.wikipedia.org/wiki/Lawsone (https://en.wikipedia.org/wiki/Lawsone)
IUPAC name 2-Hydroxy-1,4-naphthoquinone
CAS Number 83-72-7
Chemical formula C10H6O3
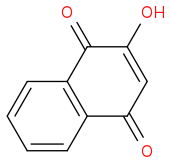
Molar mass 174.15 g/mol
Citric acid
https://en.wikipedia.org/wiki/Citric_acid (https://en.wikipedia.org/wiki/Citric_acid)
Preferred IUPAC name 2-Hydroxypropane-1,2,3-tricarboxylic acid
CAS Number 77-92-9
Chemical formula C6H8O7
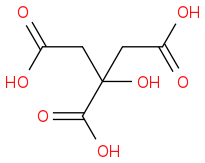
Molar mass 192.12 g/mol
Post by: billnotgatez on October 29, 2017, 09:26:44 PM
https://en.wikipedia.org/wiki/Benzoic_acid (https://en.wikipedia.org/wiki/Benzoic_acid)
Systematic IUPAC name Benzenecarboxylic acid
CAS Number 65-85-0
Chemical formula C7H6O2 or C6H5COOH
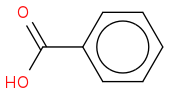
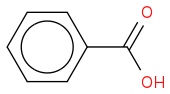
Molar mass 122.12 g/mol
Melting point 122.41 C (252.34 F; 395.56 K)
Post by: Borek on November 07, 2017, 02:56:17 AM
![C([C@@H]1[C@H]([C@@H]([C@H]([C@H](O1)O)O)O)O)O](https://www.chemicalforums.com/SMILES/d02e3e8504eb96b957c7.png)
![OC[C@H]1OC(O)[C@H](O)[C@@H](O)[C@@H]1O](https://www.chemicalforums.com/SMILES/10cad4d5f70df7a5d74f.png)
Post by: billnotgatez on March 18, 2018, 03:21:24 AM
IUPAC name (1R,2S,3r,4R,5S,6s)-cyclohexane-1,2,3,4,5,6-hexayl hexakis[dihydrogen (phosphate)]
https://en.wikipedia.org/wiki/Phytic_acid
CAS Number 83-86-3
Chemical formula C6H18O24P6
![[C@@H]1([C@@H]([C@@H]([C@@H]([C@H]([C@@H]1OP(=O)(O)O)OP(=O)(O)O)OP(=O)(O)O)OP(=O)(O)O)OP(=O)(O)O)OP(=O)(O)O](https://www.chemicalforums.com/SMILES/a3077b3f6218f9066f2e.png)
Molar mass 660.03 g/mol
Post by: Elric82 on April 04, 2018, 06:00:35 PM
Just a test on my message formatting
After all, that’s what this thread is for, isn’t it? I think this thread should be a sticky, but hey, I’m too knew around here to make such suggestions. Even though I’ve lurked here for a long time. Just a thought.
Post by: Borek on April 04, 2018, 06:12:36 PM
I think this thread should be a sticky
Good idea.
Post by: sjb on April 05, 2018, 05:17:34 AM
Good idea.
I thought it was at one stage, maybe things were broken on an update.
Post by: Borek on April 05, 2018, 06:46:07 AM
I thought it was at one stage, maybe things were broken on an update.
Or someone (could be me) unstickied it accidentally at some point.
Post by: billnotgatez on July 23, 2018, 01:52:04 PM
the above was create using url rather than zrl below
[zrl=https://en.wikipedia.org/wiki/Clausius%E2%80%93Clapeyron_relation]Clausius–Clapeyron relation[/zrl]
Post by: billnotgatez on July 29, 2018, 02:56:57 PM
Mass Relationships of Simple Compounds of Nitrogen and Oxygen
Compound Total Mass (g) Mass of Nitrogen (g) Mass of Oxygen (g)
Oxide A 3.28 1.00 2.28
Oxide B 2.14 1.00 1.14
Oxide C 1.57 1.00 0.57
Possible Molecular Formulae for Nitrogen Oxides
Assuming that:
Oxide C is NO Oxide B is NO Oxide A is NO
Oxide A is NO4 NO2 NO
Oxide B is NO2 NO N2O
Oxide C is NO N2O N4O
Post by: billnotgatez on September 09, 2018, 12:52:09 PM
![[Na+].[Na+].[O-]B1OB2OB([O-])OB(O1)O2.O.O.O.O.O.O.O.O.O.O](https://www.chemicalforums.com/SMILES/fded39fac63eee863776.png)
Post by: AWK on May 19, 2019, 11:03:00 AM
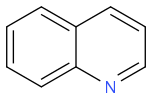
C1=CC=C2C(=C1)C=CC=N2
Post by: billnotgatez on December 07, 2019, 09:54:34 AM
![O[C@H]1C[C@H]2CC[C@]1(C)C2(C)C](https://www.chemicalforums.com/SMILES/e00ffc82d718a3bee4ac.png)
https://en.wikipedia.org/wiki/Borneol
Post by: billnotgatez on January 24, 2020, 06:54:03 AM
testing spell checker
shows misspelled for me
Post by: billnotgatez on December 09, 2020, 01:19:03 PM
Cu(ClO3)2 (aq)
NH4ClO3 (aq)
CuI (s)
Post by: Corribus on December 10, 2020, 06:45:27 PM
The following site has a nice sandbox for testing out LaTeX code (and is a good reference as well).I used this all the time and the sandbox doesn't seem to be there any more. Am I the only one with this problem?
http://www.forkosh.com/mimetextutorial.html
In the meantime, will use this space for some testing Tex code
[tex]C=kt \frac{L-x}{L}-\frac{kL^2}{D}\left(\frac{x}{3L}-\frac{x^2}{2L^2}+\frac{x^3}{6L^3}\right)+\frac{2kL^2}{D\pi^3}\sum_{n=1}^{\infty}\frac{1}{n^3}\sin{\left( \frac{n \pi x}{L} \right)}\exp{\left( -\frac{Dn^2 \pi^2 t}{L^2}\right)}[/tex]
[tex]C=kt \frac{L-x}{L}+k\frac{(L-x)^3}{8LD}+\frac{2kL^2}{D\pi^3}\sum_{n=1}^{\infty}\frac{1}{n^3}\sin{\left( \frac{n \pi x}{L} \right)}\exp{\left( -\frac{Dn^2 \pi^2 t}{L^2}\right)}[/tex]
Post by: billnotgatez on December 14, 2020, 07:47:44 PM
[tex] Situation 2 --> (R) = \frac{(P_2 . V_2)}{(n_2 . T_2) } [/tex]
Post by: Borek on December 15, 2020, 03:35:46 AM
[tex] Situation 1 --> (R) = \frac{(P_1 . V_1)}{(n_1 . T_1) } [/tex]
[tex]\rightarrow~~\longrightarrow~~\Rightarrow~~\Longrightarrow[/tex]
Post by: billnotgatez on December 15, 2020, 04:43:46 AM
Post by: billnotgatez on December 15, 2020, 07:28:11 AM
[tex] Situation 2 \Longrightarrow~~ (R) = \frac{(P_2 . V_2)}{(n_2 . T_2) } [/tex]
Post by: AWK on December 15, 2020, 07:39:56 AM
[tex] Situation 1 \Longrightarrow~~ (R) = \frac{(P_1 \cdot V_1)}{(n_1 \cdot T_1) } [/tex]
[tex] Situation 2 \longrightarrow~~ (R) = \frac{(P_2 \cdot V_2)}{(n_2 \cdot T_2) } [/tex]
Post by: billnotgatez on December 22, 2020, 02:18:47 AM
[tex] Situation 2 ~ \Longrightarrow ~~ (R) = \frac{(P_2 \cdot V_2)}{(n_2 \cdot T_2) } [/tex]
checking to see which multiplication symbol I like best
Post by: Arkcon on January 13, 2021, 07:59:53 AM
Humph. Seems not to work for me. Chiral methanol? Anyone? That's where two of the hydrogens are replaced with a tritium and a deuterium.
Post by: AWK on January 13, 2021, 08:26:07 AM
![C(O)([2H])([H])[3H]](https://www.chemicalforums.com/SMILES/d3ea7c7d5665a22e3a8d.png)
Post by: billnotgatez on January 17, 2021, 09:03:32 PM
4-Nitrobenzoic acid
Other names
p-Nitrobenzoic acid
Chemical formula
C7H5NO4
c1ccc(C(=O)O)cc1](https://www.chemicalforums.com/SMILES/46266a6e0c52a564875b.png)
https://en.wikipedia.org/wiki/4-Nitrobenzoic_acid (https://en.wikipedia.org/wiki/4-Nitrobenzoic_acid)
IUPAC name
3-Nitrobenzoic acid
Other names
m-Nitrobenzoic acid
Chemical formula
C7H5NO4
c1cc(C(=O)O)ccc1](https://www.chemicalforums.com/SMILES/9e2d23781edcb3572aef.png)
https://en.wikipedia.org/wiki/3-Nitrobenzoic_acid (https://en.wikipedia.org/wiki/3-Nitrobenzoic_acid)
IUPAC name
2-Nitrobenzoic acid
Other names
o-Nitrobenzoic acid
Chemical formula
C7H5NO4
c1c(C(=O)O)cccc1](https://www.chemicalforums.com/SMILES/5bc4bbcfa1b283bb9d96.png)
https://en.wikipedia.org/wiki/2-Nitrobenzoic_acid (https://en.wikipedia.org/wiki/2-Nitrobenzoic_acid)
Post by: billnotgatez on June 22, 2021, 09:30:20 AM
https://en.wikipedia.org/wiki/Oxytocin (https://en.wikipedia.org/wiki/Oxytocin)
[C@@H]1NC(=O)[C@H](Cc2ccc(O)cc2)NC(=O)[C@@H](N)CSSC[C@H](NC(=O)[C@H](CC(N)=O)NC(=O)[C@H](CCC(N)=O)NC1=O)C(=O)N3CCC[C@H]3C(=O)N[C@@H](CC(C)C)C(=O)NCC(N)=O](https://www.chemicalforums.com/SMILES/af980cc911e4090d9ae5.png)
Post by: billnotgatez on July 24, 2021, 09:13:23 AM
Chemical formula CH2Cl2
Molar mass 84.93 g·mol−1
Density 1.3266 g/cm3 (20 °C)
Melting point −96.7 °C (−142.1 °F; 176.5 K)
Boiling point 39.6 °C (103.3 °F; 312.8 K)
https://en.wikipedia.org/wiki/Dichloromethane
Bromine, 35Br
Standard atomic weight Ar, std(Br) [79.901, 79.907] conventional: 79.904
Atomic number (Z)35
Phase at STP liquid
Melting point(Br2) 265.8 K (−7.2 °C, 19 °F)
Boiling point(Br2) 332.0 K (58.8 °C, 137.8 °F)
Density (near r.t.)Br2, liquid: 3.1028 g/cm3
https://en.wikipedia.org/wiki/Bromine
Post by: billnotgatez on August 18, 2021, 10:11:10 AM
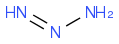
https://en.wikipedia.org/wiki/Tetrazene (https://en.wikipedia.org/wiki/Tetrazene)
edit later for error
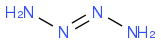 :larrow: correct
:larrow: correctsee above
Post by: sjb on August 18, 2021, 12:18:51 PM
Post by: billnotgatez on August 18, 2021, 01:27:22 PM
thanks for catching my error
Post by: billnotgatez on August 24, 2021, 12:46:13 AM
1.008 * 10 = 10.08
10.81 * 4 = 43.24
15.999 * 12 = 191.988
22.98976928 * 2 = 45.97953856
291.28753856
Post by: billnotgatez on March 26, 2022, 04:44:43 PM
2NaOH(aq)+H2SO4(aq)→2H2O(l)+Na2SO4(aq)
formatted
Post by: billnotgatez on March 26, 2022, 04:49:30 PM
2NaOH(aq)+H2SO4(aq)→2H2O(l)+Na2SO4(aq)
copy paste
Post by: billnotgatez on March 26, 2022, 04:50:56 PM
H2SO4 + NaOH = Na2SO4 + H2O
2NaOH(aq)+H2SO4(aq)→2H2O(l)+Na2SO4(aq)
quoted
Post by: billnotgatez on March 26, 2022, 05:10:19 PM
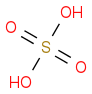
NaOH
![[OH-].[Na+]](https://www.chemicalforums.com/SMILES/de802dc49e59bd7d0a60.png)
Na2SO4
![[Na+].[Na+].[O-]S([O-])(=O)=O](https://www.chemicalforums.com/SMILES/e7e583164e59acbe0894.png)
H2O

not HOH oh well?
using smiles
Post by: billnotgatez on April 17, 2022, 06:54:24 PM
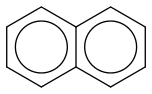
Naphthalene
https://en.wikipedia.org/wiki/Naphthalene (https://en.wikipedia.org/wiki/Naphthalene)
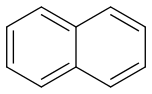
Naphthalene
https://pubchem.ncbi.nlm.nih.gov/#query=naphthalene (https://pubchem.ncbi.nlm.nih.gov/#query=naphthalene)
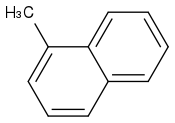
1-Methylnaphthalene
https://pubchem.ncbi.nlm.nih.gov/compound/1-methylnaphthalene (https://pubchem.ncbi.nlm.nih.gov/compound/1-methylnaphthalene)
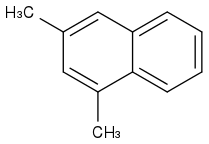
1,3-Dimethylnaphthalene
https://pubchem.ncbi.nlm.nih.gov/compound/1_3-Dimethylnaphthalene (https://pubchem.ncbi.nlm.nih.gov/compound/1_3-Dimethylnaphthalene)
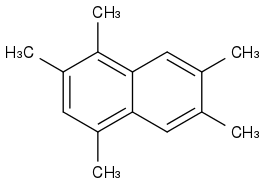
1,2,4,6,7-pentamethylnaphthalene
https://pubchem.ncbi.nlm.nih.gov/#query=1%2C2%2C4%2C6%2C7-pentamethylNaphthalene (https://pubchem.ncbi.nlm.nih.gov/#query=1%2C2%2C4%2C6%2C7-pentamethylNaphthalene)
Post by: billnotgatez on April 27, 2022, 08:50:08 PM
Post by: billnotgatez on May 05, 2022, 08:55:21 AM
https://en.wikipedia.org/wiki/Citric_acid (https://en.wikipedia.org/wiki/Citric_acid)
UPAC name
Citric acid
Preferred IUPAC name
2-Hydroxypropane-1,2,3-tricarboxylic acid
Chemical formula
C6H8O7
Molar mass
192.123 g/mol (anhydrous),
210.14 g/mol (monohydrate)

Post by: billnotgatez on May 05, 2022, 12:51:05 PM
Methyl orange
https://en.wikipedia.org/wiki/Methyl_orange
Preferred IUPAC name
Sodium 4-{[4-(dimethylamino)phenyl]diazenyl}benzene-1-sulfonate
Other names
Sodium 4-[(4-dimethylamino)phenylazo]benzenesulfonate
CAS Number
547-58-0
Chemical formula
C14H14N3NaO3S
Molar mass
327.33 g·mol−1
![[Na+].CN(C)c2ccc(/N=N/c1ccc(cc1)S([O-])(=O)=O)cc2](https://www.chemicalforums.com/SMILES/c164c487e33e657415db.png)
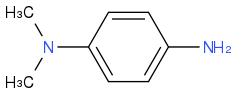
Dimethyl-4-phenylenediamine
https://en.wikipedia.org/wiki/Dimethyl-4-phenylenediamine (https://en.wikipedia.org/wiki/Dimethyl-4-phenylenediamine)
Preferred IUPAC name
N1,N1-Dimethylbenzene-1,4-diamine
Other names
p-Aminodimethylaniline;
N,N-Dimethyl-p-phenylenediamine;
4-(Dimethylamino)aniline;
p-Amino-N,N-dimethylaniline;
p-(Dimethylamino)aniline;
DMPPDA;
Dimethyl-p-phenylenediamine;
4-Amino-N,N-dimethylaniline;
p-Dimethylaminophenylamine;
DMPD
CAS Number
99-98-9
Chemical formula
C8H12N2
Molar mass
136.198 g·mol−1
Post by: billnotgatez on May 23, 2022, 10:10:38 AM
![C1=CC(=CC=C1N)S(=O)(=O)[O-].O.[Na+]](https://www.chemicalforums.com/SMILES/5144b41b4959e23fa760.png)
sodium;4-aminobenzenesulfonate;hydrate
Post by: billnotgatez on September 23, 2022, 09:45:51 PM
Ca(CN)2
Calcium cyanamide
CaCN2
Post by: Borek on October 20, 2022, 12:21:19 PM
Post by: billnotgatez on December 16, 2022, 01:49:47 PM
Chemical formula C6H5ClO2S
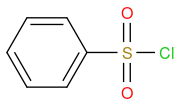
Post by: billnotgatez on March 30, 2023, 08:46:00 PM
1 Standard atmosphere = 101.325 Kilopascal
1 Standard atmosphere = 101325 Pascal
1 Standard atmosphere = 14.6959 Pound force per square inch
1 Standard atmosphere = 760 Torr
Post by: billnotgatez on June 14, 2023, 12:16:24 AM
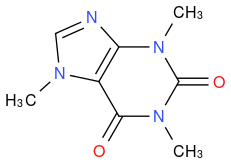
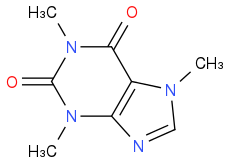
Caffeine
Post by: billnotgatez on February 23, 2024, 12:38:40 AM
![[Na+].[O-]C(=O)c1ccccc1](https://www.chemicalforums.com/SMILES/fe7532eef48c7e24c5d0.png)
Vitamin C
[C@H]1OC(=O)C(O)=C1O](https://www.chemicalforums.com/SMILES/40bffdc354a2337d1c22.png)
Benzene

![C(C)(C)[C@@H]1CC[C@H](C1)O](https://www.chemicalforums.com/SMILES/f41365db29d2964ba540.png)