though I don't know little about the fines of biochemistry involved here , first thing that comes to mind is kinetics: if the reaction with your crosslinker follows third order kinetics - i.e. the concentrations of components A , B to be crosslinked and the crosslinker C would act like this:
v = k * [A] * [B ] * [C]
(in case of A = B hence v = k * [A]
2*[C] )
it follows that, if you reduce [A], [B ] and [C] by factor of 5 each, you'd need 5
3 = 125 times more time to achieve the same result , compared to undiluted reaction kinetics.
30 minutes at 10 mg/mL
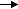
2.6 days at 2 mg/mL
just ballpark considerations, but in general, something like that might happen and turn out to be a problem, if you didn't allow for much longer reaction time than in your original protocol
regards
Ingo