please tell me Why do we need both Nmr's ? iam planning to get only 1H nmr ? isn't it alone sufficient ?
While a
1H NMR is often sufficient for a quick identification of simple molecules with good confidence, it doesn't provide information about the C atoms, so it is not enough for a completely convincing structural assignment -
13C NMR is more evidence for the assigned structure, the more evidence you have, the more confidence in your assignment. In some cases, even more NMR experiments would be required for convincing assignment.
A simple case illustrating the need for
13C is:
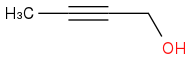
and
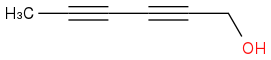
They would have very similar
1H NMR spectra.
Many journals insist on
13C spectra for novel compounds. Whether
1H is enough for publication depends on the molecule, the method of preparation and the journal in which it is published. My personal opinion is that you should report a
13C spectrum for any novel compound regardless of whether the journal requires it.
If the compound is not novel - i.e. it has already been characterized in the literature - then
1H NMR is "enough" provided your data matches the literature, and the literature interpretation of the data is sound.